esiRNA coding mouse
The esiRNA technology is a powerful tool for loss of function gene studies. To illustrate the potency of silencing triggers quantitative real time (qRT)-PCR is often used to measure the knock-down rates. The knock-down validation on mRNA level was here performed 24 hours post transfection of esiRNA using qRT-PCR. To be able to assess knock-down rates, the expression levels of the mRNA of interest was compared to cells (HeLa or mouse ES) simultaneously transfected with Renilla Luciferase (negative control).
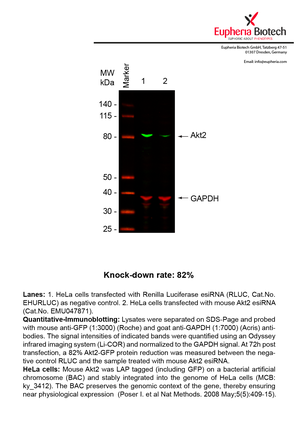
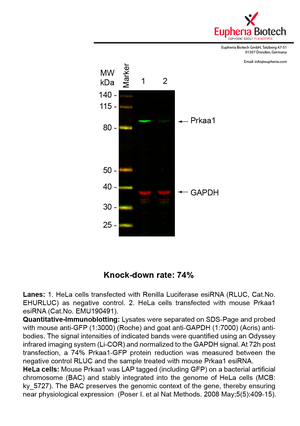
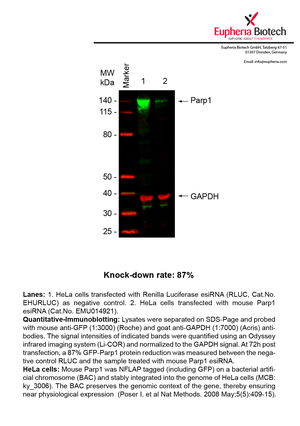
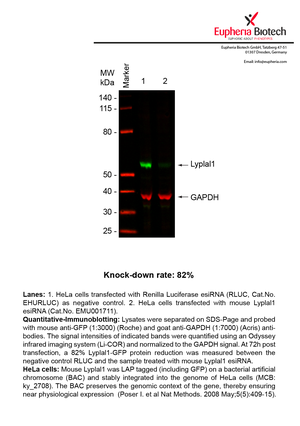
However, a more valuable measure of the knock-down potency in an RNAi experiment is the reduction in protein level. We therefore also validate the knock-down rates of esiRNAs on protein level using quantitative western blot analysis (Odyssey, Li-COR). The time point for maximum knock-down rate for each protein can vary significantly, as it is depending on factors such as protein-stability, turn-over rate or cell proliferation rates. The knock-down validation on protein level was here performed at 72 hours post transfection of esiRNA in HeLa cells. To be able to assess knock-down rates, the expression levels of the protein of interest was compared to HeLa cells simultaneously transfected with Renilla Luciferase (negative control).
Two approaches were used to monitor knock-down at protein level: 1. Specific antibodies for the protein of interest were used for the quantitative western blot analysis. 2. Proteins of interest were GFP-tagged on a bacterial artificial chromosome (BAC) and stably integrated into the genome of HeLa cells, allowing for near physiological expression (Poser I. et al Nat Methods. 2008 May;5(5):409-15). Using a GFP antibody the detection by quantitative western blot analysis of the protein of interest is straightforward.
| esiRNA ID | Gene Name | Gene Description | Ensembl ID | RefSeq ID | Knock-Down Rate | Pic / PDF |
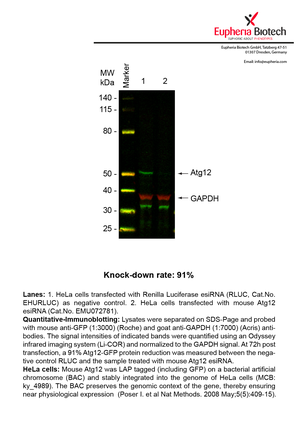
| EMU072781 | Atg12 | autophagy-related 12 (yeast) | ENSMUSG00000032905 | NM_026217 | 91% | Pic / PDF |
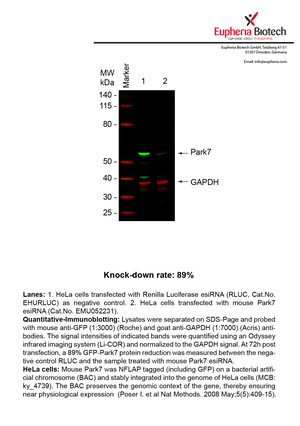
| EMU052231 | Park7 | Parkinson disease (autosomal recessive, early onset) 7 | ENSMUSG00000028964 | NM_020569 | 89% | Pic / PDF |
| EMU014921 | Parp1 | poly (ADP-ribose) polymerase family, member 1 | ENSMUSG00000026496 | NM_007415 | 87% | Pic / PDF |
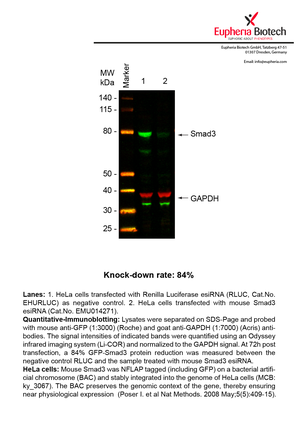
| EMU014271 | Smad3 | MAD homolog 3 (Drosophila) | ENSMUSG00000032402 | NM_016769 | 84% | Pic / PDF |
| EMU047871 | Akt2 | thymoma viral proto-oncogene 2 | ENSMUSG00000004056 | NM_001110208 | 82% | Pic / PDF |
| EMU001711 | Lyplal1 | lysophospholipase-like 1 | ENSMUSG00000039246 | NM_146106 | 82% | Pic / PDF |
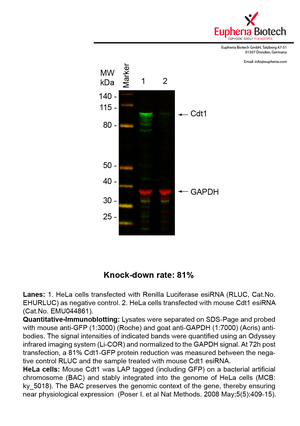
| EMU044861 | Cdt1 | chromatin licensing and DNA replication factor 1 | ENSMUSG00000006585 | NM_026014 | 81% | Pic / PDF |
| EMU190491 | Prkaa1 | protein kinase, AMP-activated, alpha 1 catalytic subunit | ENSMUSG00000050697 | 74% | Pic / PDF |